Developmental epigenetics and the role of DNA methylation in reproductive health
What role does DNA methylation play in developmental biology and genomic imprinting?
DNA methylation is a fundamental epigenetic mechanism that governs gene expression during embryonic development, cell fate determination, and genomic imprinting. Proper establishment and maintenance of methylation patterns are essential for normal growth and differentiation, while disruptions can lead to developmental abnormalities or imprinting disorders such as Prader–Willi, Angelman, and Beckwith–Wiedemann syndromes.
In developmental and reproductive research, precise methylation analysis enables scientists to uncover how epigenetic regulation shapes embryogenesis, how stem cells maintain pluripotency or commit to specific lineages, and how parental environments influence the next generation. However, these studies often rely on extremely limited or delicate DNA samples, such as blastocysts, or early embryonic tissues, that are highly sensitive to degradation during standard bisulfite treatment.
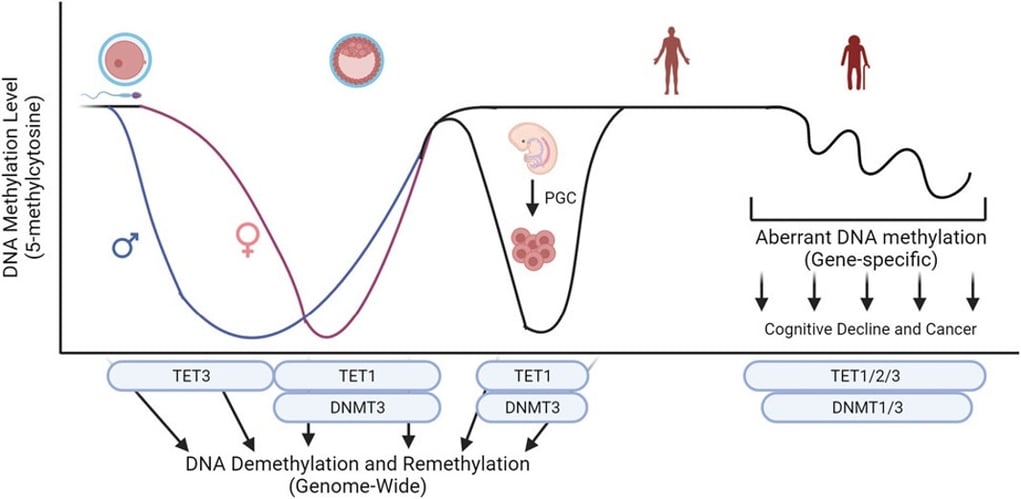
Applications for methylation analysis in developmental and genetic imprinting research
- Stem Cell Research: Study methylation reprogramming in pluripotent stem cells (iPSCs) and differentiation pathways.
- Epigenetic Inheritance Studies: Explore how parental environmental exposures influence the methylation landscape of offspring.
- Imprinting Disorder Diagnosis: Identify methylation abnormalities linked to Prader–Willi, Angelman, and Beckwith–Wiedemann syndromes.
These applications bridge basic biology and translational medicine, offering insights into epigenetic regulation across generations.
How Ellis Bio’s next generation bisulfite chemistry enables advanced developmental epigenetic research
Ellis Bio’s next-generation (Ultra-Mild and Ultra-Fast) bisulfite chemistry is built to meet the unique demands of developmental and imprinting research, where DNA samples are often low in quantity and highly sensitive.
-
Precision in Imprinting Analysis: Complete and sequence bias-free C-to-T conversion allows reliable detection of parent-of-origin–specific methylation patterns critical for studying genomic imprinting and epigenetic inheritance.
- Consistency for Developmental Studies: Reproducible C-to-T conversion and High-yield recovery enable confident comparisons across developmental stages and differentiation experiments.
- Seamless Integration with Automation Workflows: Simple and fast workflow fully compatible with NGS and DNA methylation microarray platforms, enabling efficient large-scale and high-throughput studies.
References
- Smith, Zachary D., Sara Hetzel, and Alexander Meissner. "DNA methylation in mammalian development and disease." Nature Reviews Genetics
- Wilkinson, Amy L., Irene Zorzan, and Peter J. Rugg-Gunn. "Epigenetic regulation of early human embryo development." Cell Stem Cell
- Greeson, Katherine W., et al. "Inheritance of paternal lifestyles and exposures through sperm DNA methylation." Nature Reviews Urology
